Application note: Size distribution analysis of influenza virus particles using the CPS disc centrifuge

Influenza virus production for seasonal vaccines
By Michael M. Pieler and Michael Wolff, Otto-von-Guericke-Universität, Magdeburg
Vaccination is an important measure to control influenza virus outbreaks. Therefore, large efforts are being undertaken to optimize current production processes and to develop new approaches for influenza virus vaccine manufacturing.
One factor with increasing importance in newly designed processes is virus particle aggregation. Depending on the employed unit operation during downstream processing (DSP), e.g. (crossflow-)filtration, chromatography or centrifugation, product losses due to aggregation can be substantial.
As a result, several process conditions are currently investigated for assessing their impact on virus particle aggregation and to get a better understanding of the involved effects.
Method description
Size distributions of influenza virus strains (A/Puerto Rico/8/34, A/Wisconsin/67/2005, A/Eq/Newmarket, and B/Malaysia/2506/2004) produced in MDCK cells were determined.1,2,3 Analysis was carried out with differential centrifugal sedimentation on a CPS DC24000 UHR disc centrifuge as described by Neumann et al.4 Measurements were performed at 24,000 rpm with a sucrose gradient ranging from 2 to 8 % w/v in 1x phosphate buffer saline (1x PBS). The gradient was equilibrated for 30 min and 200 µL sample was injected per analysis. Measurement time per sample was about 15 min. Each sample is represented in the following figures as an average of three technical replicates in new gradients.
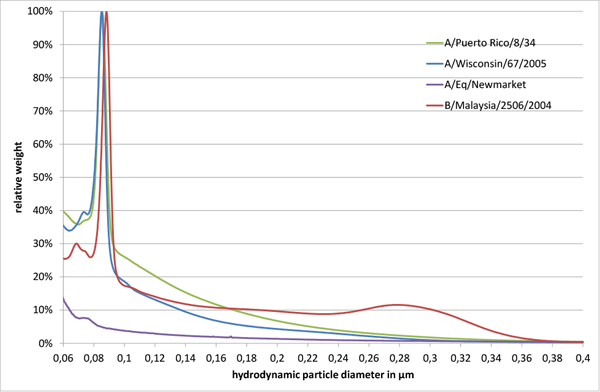
Fig. 1
Results and discussion
All tested influenza virus strains showed a major virus peak in the size range of 80 to 90 nm. For B/Malaysia/2506/2004 an additional broader peak was observed around 280 nm (Fig. 1). Furthermore, small peaks around 70 to 80 nm were observed, which might be attributed to cell debris or virus fragments.
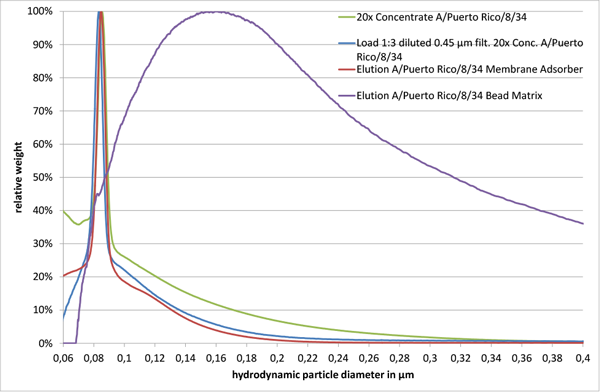
Fig. 2
The size distribution of a 20x concentrated A/Puerto Rico/8/34 sample after a 1:3 dilution in 10 mM TRIS HCl pH 7.4 and a 0.45 µm filtration showed only minor differences, when compared to the initial concentrated virus sample (“Load 1:3 diluted 0.45 µm filt. 20x Conc. A/Puerto Rico/8/34” and “20x Concentrate A/Puerto Rico/8/34” in Fig. 2). However, analysis of eluted virus particles from the membrane adsorber and the bead chromatographic matrix with equivalent functional groups and similar backbone structure resulted in different particle size distributions. Virus particles eluted from the membrane adsorber (“Elution A/Puerto Rico/8/34 Membrane Adsorber” in Fig. 2) revealed a sharp major peak at 85 nm, alike the material loaded onto it (“Load 1:3 diluted 0.45 µm filt. 20x Conc. A/Puerto Rico/8/34” and “20x Concentrate A/Puerto Rico/8/34” in Fig. 2). However, the corresponding product fraction of the bead matrix showed a broad peak ranging from approximately 80 nm up to 2 µm (“Elution A/Puerto Rico/8/34 Bead Matrix” in Fig. 2, size range only shown up to 400 nm).
Conclusion
Minor changes in process conditions can potentially lead to significant variations in particle size distributions of influenza virus particles, which can result in substantial reductions in product yield and purity. An increasing attention to virus aggregation can drive the design of new vaccine manufacturing processes and improve their overall performance and robustness. For this purpose, the CPS DC24000 UHR disc centrifuge represents a useful tool. However, analysis of virus aggregation is complex and a combination of different methods is beneficial to show the complete picture.
References
- Genzel, Y., Behrendt, I., König, S., Sann, H. & Reichl, U. Metabolism of MDCK cells during cell growth and influenza virus production in large-scale microcarrier culture. Vaccine 22, 2202–2208 (2004).
- Genzel, Y., Dietzsch, C., Rapp, E., Schwarzer, J. & Reichl, U. MDCK and Vero cells for influenza virus vaccine production: a one-to-one comparison up to lab-scale bioreactor cultivation. Appl. Microbiol. Biotechnol. 88, 461–475 (2010).
- Genzel, Y., Fischer, M. & Reichl, U. Serum-free influenza virus production avoiding washing steps and medium exchange in large-scale microcarrier culture. Vaccine 24, 3261–3272 (2006).
- Neumann, A. et al. New method for density determination of nanoparticles using a CPS disc centrifugeTM. Colloids Surf. B Biointerfaces 104, 27–31 (2013).



